SpinCore RadioProcessor NMR Interface for MATLAB
Contents
- Legal
- Installation
- Single
Pulse NMR
- CPMG
NMR
- 90
Degree Pulse
Width Finder
- CYCLOPS
NMR
- Included
Functions
- Creating
Additional MATLAB Functions
Legal
Copyright (c) 2008 SpinCore Technologies, Inc.
This software is provided 'as-is', without any expressed
or implied
warranty. In no event will the authors be held liable for any damages
arising from the use of this software.
Permission is granted to anyone to use this software for
any
purpose, including commercial applications.
Installation
To install and ready the console for use, simply extract
the zip
file to the desired location. In MATLAB, change the current directory
to the location where the files were unzipped.
After changing the current directory in MATLAB, execute
Single Pulse
NMR m-file (spincore_nmr_gui.m) by either typing "spincore_nmr_gui" in
the MATLAB command window, right-clicking the file from the command
interface and choosing Run, or
running the file from MATLAB's m-file editor.
The CPMG interface (spincore_cpmg_gui.m) and 90
Degree Pulse Width Interface (spincore_pulse_width_gui.m) may be
executed
in a
similar manner.
Single Pulse NMR
This program is first and foremost an easy-to-use
version of the
single_pulse_nmr outlined in the RadioProcessor manual. Since operation
of this interface relies heavily on the original single_pulse_nmr code,
the section of the RadioProcessor manual relating to NMR is still
relevant here. Operation of the Single Pulse NMR program is dominated
by the use of a set of easily-modified input parameters and two buttons.
Performing a Scan
Upon selecting the desired input parameters and pressing
the Perform
Scan button, the parameters will be passed through MATLAB to
the RadioProcessor board to initiate the scan. Since the board can
perform the scan independent of any further user interaction, control
is returned to MATLAB and the user immediately after sending the
command to begin scanning.
It is worth noting that before beginning a scan, the
user can choose
between 2 pre-programmed pulse shapes in addition to being able to
change a wide range of other input parameters. The shape of these
pulses is available in Appendix II of the RadioProcessor manual. Note
that choosing an amplitude value of 1 produces a 4 V peak-to-peak pulse
in the desired shape.
Stopping a Scan
In the event a scan has been initiated and the user does not wish for
it to continue on in its entirety, the scan may be stopped
prematurely. To stop a scan in progress, the user should use the Stop Scan button.
Downloading Data
To view the data acquired by the board, the Download
Data
button must be pressed. Because of the RadioProcessor's design, data
can be downloaded at any time, even while the RadioProcessor is
performing a scan and acquiring data. Once the button is pressed, all
data stored in the RadioProcessor's RAM will be downloaded into MATLAB
and plotted in the desired format.
In addition to downloading the data to MATLAB, the Download
Data button also creates ascii, felix, and jcamp-dx 5.0 files
containing the data. These files are saved under the name specified in
the "filename" field at the time the button is pressed and stored in a
named subfolder in the LogFiles subdirectory
of SpnmrR3.
If the Download Data button is pressed
while a
scan is in progress, the data will still be downloaded and displayed.
In addition, a status update will appear notifying the user to the
current progress of the scan. It is important to note that this does
not interrupt or stop an active scan.
Changing the values between running the scan and
downloading the data will not impact the proper display or
storage of the data.
Retrieving Data Later
Each time data is downloaded from the RadioProcessor, a MAT-file is
created and placed in a named
folder in the LogFiles subdirectory
of SpnmrR3. To
accomodate for a variety of programs, the data is saved as a variety of
filetypes including Ascii, Felix,
J-Camp Dx, and MAT-File.
The MAT-file is designed for use in MATLAB and contains all relevant
parameters and data acquired during the scan. The MAT-File can be
viewed later in the Single Pulse NMR Interface by selecting the Load Data File option under the File menu. Upon selecting
this option, the user will be prompted to select a Mat-File associated
with the Single Pulse NMR Interface. If the user navigates to the
LogFiles
subdirectory and then to a MAT-File created in a previous session, all
the relevant data will be loaded into the interface as though the user
had just downloaded the data from the RadioProcessor.
Saving and Loading Settings
The Single Pulse NMR comes pre-loaded with a set of
default
parameters that appear everytime the program is run. In the event the
user changes these parameters and wishes to have quick access to them
later, a Save Settings option is included under the File menu.
Choosing this option will allow the user to save the current settings
as a MAT file.
Similar to the Save Settings option is
the Load
Settings option which is also located under the File menu. This option will prompt
the user to select a MAT file in which previously selected settings
have already been stored.
This feature differs from the Load Data File option in that these
functions save and load only the input parameters. The Load Data File option loads not only
the input parameters but also the generated NMR data as well.
Detecting Resonance Frequency
One of the more valuable features of the Single Pulse
NMR Interface
is its ability to automatically detect the resonant spectrometer
frequency. Available under the Edit
menu, Set to
Resonance
will run a scan, download and analyze the data to automatically
determine the resonant spectrometer frequency. Upon completion,
the Spectrometer Frequency box in the Single Pulse NMR Interface will
automatically be set to this new frequency.
The algorithm for detecting resonant frequency is not
particularly robust and therefor will not work in every scenario.
Primarily, the algorithm works by observing the power spectral density
of a given FID and observing the frequency offset of the largest
peak. The algorithm then adjusts the spectrometer frequency such
that this peak is centered at 0 Hz. In particularly noisy systems
or systems with some types of systematic errors, the resonant frequency
detection may not function properly.
Calculating Signal-to-Noise Ratio (SNR)
After performing a scan and downloading the data to the
interface,
the Calculate SNR option
becomes available under the Edit
menu. Choosing this option opens a new window which displays a
semilog plot of the FFT of the data.
To use this interface, the user must click the Select signal and noise
button. The user should then select the signal region of the data
containing the signal peak of interest. Next the user will be
prompted to select a representative section of noise from the
plot. After these steps, the plot will have a signal portion
highlighted in green and a noise portion highlighted in red. The
user can then calculate the Signal-to-Noise Ratio of the data by
pressing the Calculate SNR
button or can select new regions of the plot by again clicking the Select signal and noise button.
Currently the Signal-to-Noise Ratio is calculated as the
ratio of
the maximal peak detected in the signal region to the RMS value of the
selected noise region. These values are currently meant for
comparison purposes only and are not intended to be used as absolute
data.
Scanning Continuously
SpinCore's Single Pulse NMR Interface also has the ability to perform
continuous scans. To scan continuously, the user should set the
input parameters then simply press the Scan
Continuously button. This will then cause the
RadioProcessor board to read the input parameters and begin
scanning. In this mode, the RadioProcessor performs a scan,
downloads the data, performs another scan and so on until the user
stops the scan.
The input parameters become locked in place while the continuous scan
is running. This requires the user to temporarily stop the scan
in order to change the parameters.
Utilizing
Undersampling Functionality
As the RadioProcessor user's manual indicates, the RadioProcessor is
capable of acquiring input signals with frequencies of interest above
the Nyquist frequency. Although aliasing may occur, the
RadioProcessor is still capable of adequately detecting and acquiring
these signals. The MATLAB interface has been coded to reflect
these capabalities.
Explanation of input parameters:
Number of Points: Specify the number of
complex
data points to capture, this should be an integer between 1 and 16384.
Spectrometer Frequency: The frequency
of the
spectrometer, in MHz
Spectral Width: The desired baseband
spectral
width, in kHz.
Pulse Duration: Duration of TX
excitation pulse, in
seconds.
Ringdown Time: Transient, or ringdown
time, in
seconds.
Repetition Delay: Delay between scans,
in seconds.
Number of Scans: Number of times to
scan and
average the data.
Output Phase: Phase of the TX output
channel, in
degrees.
Clock Frequency: Clock frequency of the
board, in
MHz.
Bypass FIR: Check to bypass the FIR
filter, leave
unchecked to use it. If this is left unchecked, the maximum useable
Spectral Width is 135 kHz.
Output Amplitude Scaling: Amplitude of
excitation
pulse, a value
between 0 and 1.
File Name: Name of the file the results
of the scan
will be saved under.
Pulse Shape: Choose either to use a
hard pulse with
the scan or a sinc pulse.
Timing Diagram
In this experiment, a single scan is performed as
follows: a single
RF pulse is applied
to the sample for a specified amount of time (Pulse time). The high
voltage induced on the sample coil is
allowed to ring down (Transient time), after which the Free Induction
Decay (FID) signal is acquired (for length
Acquisition time). The sample is then allowed to relax for time
Repetition delay. This scan procedure is then
repeated an arbitrary number of times as desired to improve the signal
to noise ratio.
In the timing diagram below, tx_enable, phase_reset, and
trigger_scan are the control lines used by
the internal PulseBlaster timing core to control the experiment.
CPMG NMR
The CPMG NMR gui is an interface new to Release 2 that allows users to
quickly and easily perform CPMG NMR experiments. Modeled after
the original Single Pulse NMR interface, users will note that the two
interfaces operate in a very similar manner. Although some of the
parameters differ and the frequency domain plot as well as the SNR
calculator have been removed, the two interfaces otherwise operate
identically.
Capturing Data
in a
Continuous Scan:
To capture data in a continuous scan, the Points per Echo parameter must be
set to
0. This will cause the RadioProcessor to trigger and begin
capturing data immediately after the transient time on the 90 degree
pulse. Once data has begun to be captured in this manner, it will
continue until the desired number of echoes have been performed.
Capturing Only the Peaks of the Data:
As an alternative to capturing data continuosly, it is possible to
capture only the peaks of each echo. For this, the user must
specify some value for Points per Echo
and Number of Echoes other
than
0. In this case, the RadioProcessor will capture the number of
points specified by Points per Echo
around the peak of each baseband signal peak. Also, the total
number of data points to be downloaded in this case is equal to the
value specified by Points per Echo multiplied
by 1 plus the value specified in Number
of
Echoes.
If this results in a value greater than 16k, the user will receive an
error message.
Downloading Data
Similar to the Single Pulse NMR Interface, the scan and download
functions have been separated for most function of the CPMG NMR
Interface. As such, the user must press the Download Data button after
performing a scan if they wish to view the results of the scan.
This action not only immediately downloads and displays the data in the
current interface, but also stores all relevant information in various
log files. Note that if the Download
Data button is pressed before the entire experiment has complete
you may obtain unwanted data. If you are performing multiple
scans, the status bar will notify the user if the experiment has
completed and if it has not completed, what percentage of the
experiment has completed when the Download
Data button is pressed.
As with the Single Pulse NMR Interface, there are 4 log files created
each time the Download Data button
is pressed. These files are saved under the LogFiles subdirectory in an
automatically generated folder with the name specified in the File Name box associated with the
last scan.
Calculating T2:
For NMR samples that produce data with singular exponential decay
during a CPMG experiment, the CPMG NMR interface can attempt to
calculate the appropriate value of T2. This process is done by
first observing a series of single maximal points collected from the
peak of
each echo. The number of echos to observe and thus the number of
datapoints can be specified by changing the Number of Echoes parameters.
Users may find that their calculations for T2 vary greatly depending on
the number of echoes they specify.
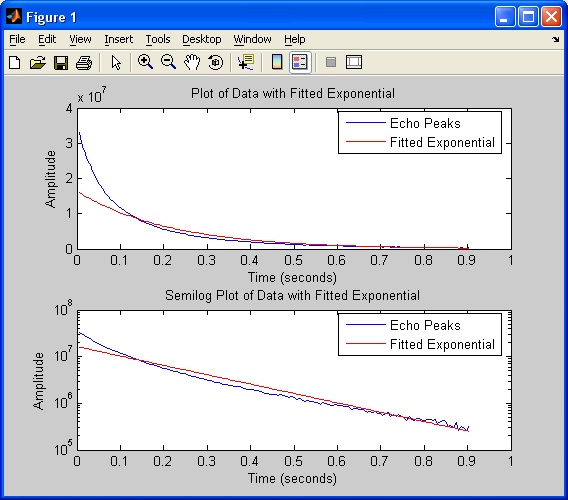
Once this data has been collected, T2 is calculated by finding the
slope of the line of best fit on a semilog plot of the peaks.
After T2 has been calculated, its value will be displayed in the
original CPMG Interface window. Also, a new figure window will be
generated containing cartesian and semilog plots of the first 150 peaks
with the calculated exponential overlayed.
Note that the T2 calculation requires that only one point per echo is
captured. If the Number of
Echoes is large, noise may dominate the signal for later echoes.
This will cause the decay to be non-exponential and the line of best
fit will not properly fit the exponential decay. For better
results, modify the number of echoes so that the acquired data, when
plotted on the semilog scale appears linear.
Explanation of Input Parameters:
Spectrometer Frequency:
The frequency
of the spectrometer, in MHz
Spectral Width: The desired baseband
spectral
width, in kHz (this value is equal to the number of points sampled per
millisecond).
Pulse Duration: Duration of 90 degree
excitation
pulse, in
microseconds. The 180 degree pulse is calculated to be twice this
value.
P2 Phase: Output phase of the 90 degree
pulse. The 180 degree pulses will have a phase offset of 90
degrees from this value.
Ringdown Time: Transient, or ringdown
time, in
microseconds.
Tau: One half
the time
between 180 degree pulses. Also, the time between the 90 degree
pulse transient time and the first 180 degree pulse.
Points per Echo: Number
of points to capture around peak of each echo. Set to 0 for
continuous scan.
Number of Echoes: Total
number of echos to capture data from. Parameter is unused during
continuous scan.
Repetition Delay: Delay between scans,
in seconds.
Number of Scans: Number of times to
scan and
average the data.
Clock Frequency: Clock frequency of the
board, in
MHz.
File Name: Name of the file the results
of the scan
will be saved under.
Bypass FIR: Check to bypass the FIR
filter, leave
unchecked to use it. If this is left unchecked, the maximum useable
Spectral Width is 135 kHz.
Timing Diagram:
90 Degree Pulse Width Finder
The 90 Degree Pulse Width Finder is a new interface designed to run a
series of single pulse scans with variable pulse widths that will
ultimately allow automatic detection of the 90 and 180 degree pulse
widths.
Pulse Width Detection
The 90 Degree Pulse Width Finder attempts to detect both 90 and 180
degree pulse widths by observing the amplitudes of the magnitude output
of a series of single pulse scans. These scans vary the pulse
width from some user-specified intial value to a user-specified final
value in user-specified increments while holding all other input
parameters constant.
After each NMR scan is completed and the FID data is acquired, an
amplitude for that FID is calculated by finding the average of a
pre-defined number of points following some offset at the beginning of
each FID scan. By default, these values are defined in the m-file
code to be 15-point averaging and a 10-point offset. Therefore,
the amplitude of each scan is taken to be the average of the 11th to
25th time-sequential amplitude values of the FID. The user is
welcomed to change these values as they please.
These calculated amplitude values are gathered for the real, imaginary
and magnitude data and are plotted sequentially in the interface until
the scan is completed or stopped. The FID that each value is
being based off of is displayed in real-time as well so that the user
may attempt to observe or diagnose any errors that may occur.
Once the full range of scans has been
completed, the software attempts to automatically detect the 90 degree
and 180 degree pulse widths using the acquired data. The software
performs this detection by observing the magnitude data amplitudes and
assigns the 90 degree pulse width to be the time index of the maximal
amplitude value.
The 180 degree pulse width is then detected as the index of the first
valley that appears in a filtered version of the magnitude data
amplitudes. This filtered version of the magnitude data is simply
the original magnitude data that has been smoothed by a simple 4-point
moving average filter in an attempt to remove any erroneous valleys
that may return improper values for the 180 degree pulse width.
Due to the somewhat simple method of detection, the pulse width values
may not be accurately calculated if the system is particularly noisy or
has significant errors.
Stopping the Scan
In the event a scan has been initiated and the user does not wish for
it to continue on in its entirety, the scan may be stopped
prematurely. To stop a scan in progress, the user should use the Stop Scan button. Once the
scan is stopped the user can still change the plot and open the figure
in a new window as though the scan had completed. The
automatic pulse width detection will not occur, however, unless a scan
is allowed to run until completion.
Explanation of Input Parameters
Number of Points:
Specify the number of
complex
data points to capture in each scan, this should be an integer between
1 and 16384.
Spectrometer Frequency: The frequency
of the spectrometer, in MHz
Spectral Width: The desired baseband
spectral
width, in kHz.
Ringdown Time: Transient, or ringdown
time, in
microseconds.
Repetition Delay: Delay or relaxation time between scans,
in seconds.
Number of Scans: Number of times to
scan and
average the data.
Output Phase: Phase of the TX output
channel, in
degrees.
A/D Clock Frequency: Clock frequency of the
board, in
MHz.
Output Amplitude Scaling: Amplitude of excitation
pulse, a value
between 0 and 1.
Bypass FIR: Check to bypass the FIR
filter, leave
unchecked to use it. If this is left unchecked, the maximum useable
Spectral Width is 135 kHz.
Pulse Shape: Choose either to use a
hard pulse with
the scan or a sinc pulse.
Intial Pulse Width: The initial pulse width to begin
scanning from.
Final Pulse Width: The final pulse width to terminate
scan at.
Step Size: The step size between pulse widths. This
can be used to improve the resolution of the detected pulse widths.
CYCLOPS NMR
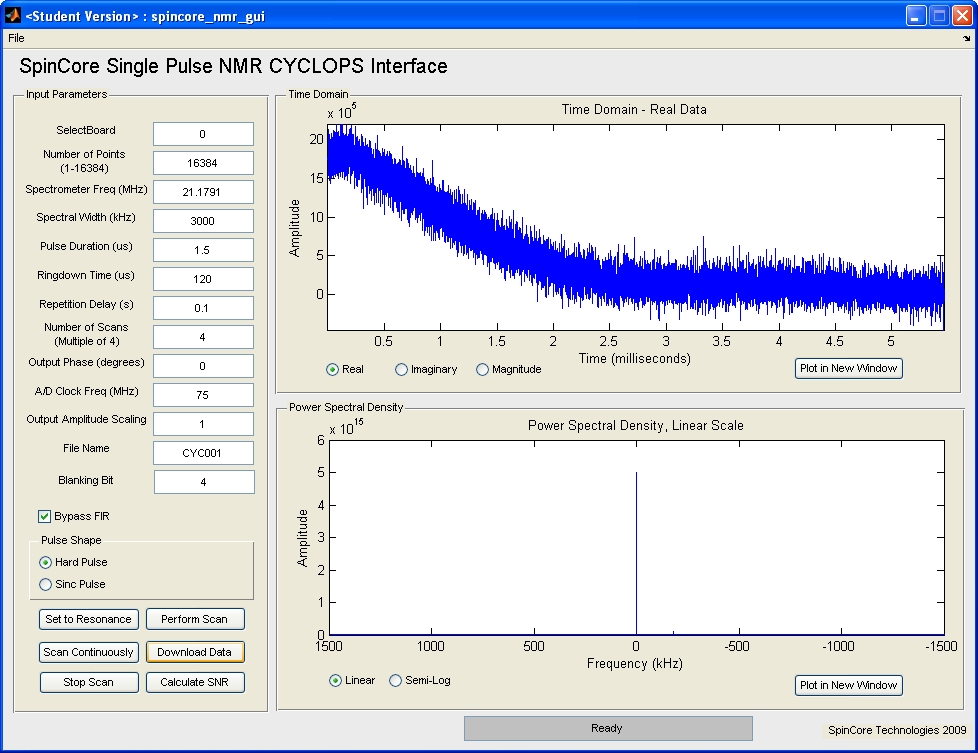
Explanation of input parameters:
Number of Points: Specify the number of
complex
data points to capture, this should be an integer between 1 and 16384.
Spectrometer Frequency: The frequency
of the
spectrometer, in MHz
Spectral Width: The desired baseband
spectral
width, in kHz.
Pulse Duration: Duration of TX
excitation pulse, in
seconds.
Ringdown Time: Transient, or ringdown
time, in
seconds.
Repetition Delay: Delay between scans,
in seconds.
Number of Scans: Number of times to
scan and
average the data. As the nature of CYCLOPS requires 4 number of scans
to complete an experiment, the number entered here must be a multiple
of 4.
Output Phase: Phase of the TX output
channel, in
degrees.
Clock Frequency: Clock frequency of the
board, in
MHz.
Bypass FIR: Check to bypass the FIR
filter, leave
unchecked to use it. If this is left unchecked, the maximum useable
Spectral Width is 135 kHz.
Output Amplitude Scaling: Amplitude of
excitation
pulse, a value
between 0 and 1.
File Name: Name of the file the results
of the scan
will be saved under.
Pulse Shape: Choose either to use a
hard pulse with
the scan or a sinc pulse.
Included MATLAB Functions
The source code for several C programs that have been
written
specially for use with MATLAB are included with SPNMR3 under the MEXSource
subdirectory.
These programs have also been
precompiled with the MATLAB mex function and can be called just like
any other function in MATLAB. It is important to note that for
these functions to work properly, MATLAB must be made aware of the
location of the function's associated dll file if it is not located in
MATLAB's current directory. The precompiled version of
these programs have the .dll extension and are located in the EssentialGuiFiles subdirectory of SPNMR3.
These functions, in their present form, provide a
powerful group of tools for interaction between the RadioProcessor and
MATLAB. Particularly, they allow for scripting and construction
of more advanced programs for the simplistic acquisition and importing
of data.
These included functions are outlined in the table below:
| Name |
Prototype |
Description |
| pbGetData |
[REAL, IMAG]=pbGetData() |
This basic function accesses the RadioProcessor,
downloads
the data from RAM and stores the real and imaginary data in the
matrices REAL and IMAG, respectively. While this is a quick and easy
way to import RadioProcessor data into MATLAB, the data lacks any
frequency information and is simply a list of magnitudes.
Using this function does not clear the data from
the RAM on
the RadioProcessor, nor does it interfere with a scan in
progress. As such, data may be acquired and displayed many times
before a longer scan has completed.
|
| pbGetDataPlus |
[REAL, IMAG, STATUS,
RESONANCE_FREQ]=pbGetDataPlus(num_points,
SF, SW,
'filename', num_scans, adc_freq, pulse_time, trans_time,
repetition_delay, tx_phase, bypass_fir, amp_val, shape); |
A more advanced version of pbGetData, this not
only stores
the data from RAM into REAL and IMAG but also creates ascii, felix, and
jcamp-dx files in the active directory using the specified filename.
This function requires additional inputs for
creating the
additional files. These inputs are the number of points, spectrometer
frequency, spectral width, filename, number of scans ,adc frequency,
pulse_time, trans_time, repetition_delay, tx_phase, bypass_fir, amp_val
and shape
respectively.
The value returned to STATUS specifies whether or
not the
scan was successful. A 0 indicates that the data was successfully read
from the RAM. For further information regarding this function, consult
the SpinAPI documentation concerning the pbGetData function.
The latest version of spinapi now supports
calculating resonance frequency if available from the data stored in
the RadioProcessor board. However this value does not gaurantee the
correct resonance frequency if there exist number of different peaks in
the data.
This function's capabilities regarding the
ability to acquire data during a scan match those of pbGetData in that
neither the RAM is cleared nor is a scan in progress stopped when this
function is called.
|
| pbReadStatus |
[STATUS, SCANS] = pbReadStatus() |
This function, when called returns the current
status of the
RadioProcessor Board as well as the current number of scans.
For information concerning the value returned to
STATUS,
consult the SpinAPI documentation concerning the pbReadStatus function.
|
| pbStop |
pbStop() |
This function stops any scan in progress. It is
not necessary
to use this function first if the user wishes to begin a new scan.
Moreover, this function is intended for users that wish to stop a scan
without beginning a new one.
|
| SpnmrScan |
SpnmrScan(num_points, SF, SW, pulse_time,
trans_time, repetition_delay, num_scans, tx_phase, bypass_fir,
adc_freq, amp_val) |
This function, when called, causes the
RadioProcessor Board
to perfrom a single pulse NMR experiment using the user-specified input
parameters. This function returns control to MATLAB immediately after
sending instructions to the RadioProcessor board and does not wait for
the scan to complete before returning control.
Calling this function does not return any outputs
to MATLAB.
Though status updates are displayed, no actual data is stored as a
result of the function call.
Data acquired during the scan is stored in RAM on
the
RadioProcessor board and may be retrieved with pbGetData or
pbGetDataPlus.
|
| SpnmrWithDelay |
SpnmrWithDelay(num_points, SF, SW, pulse_time,
trans_time,
repetition_delay, num_scans, tx_phase, bypass_fir, adc_freq, amp_val) |
This function operates identically to
SinglePulseShapeScan
with the exception that the function does not return control to MATLAB
until the scan has completed.
This function is especially useful for use in
writing scripts
that require multiple scans to be run consecutively.
|
CpmgScan
|
CpmgScan(SF, SW,
P2_time,
P2_phase, P2_after, tau, echo_points, echo_loops, repetition_delay,
num_scans, bypass_fir, adc_freq)
|
This function, when
called,
signals the RadioProcessor Board to perform a CPMG NMR experiment using
the provided input parameters. Similar to SinglePulseShapeScan,
control is returned to MATLAB immediately after the instructions are
sent to the RadioProcessor Board.
Also similar to SinglePulseShapeScan, CpmgScan does not return any data
to MATLAB. Data must be acquired using either pbGetData or
pbGetDataPlus.
|
CyclopsScan
|
[ERROR] =
CyclopsScan(num_points, SF, SW, pulse_time, trans_time,
repetition_delay, num_scans, tx_phase, bypass_fir, adc_freq, amp_val,
shape, blanking_Bits, num_Boards);
|
This function, when
called, causes the
RadioProcessor Board
to perfrom number of CYCLOPS NMR experiments specified in 'num_scans'
using the user-specified input
parameters. This function returns control to MATLAB immediately after
sending instructions to the RadioProcessor board and does not wait for
the scan to complete before returning control.
Returns:
0 is returend on success. otherwise the value has following
meanings
-1 : The board does not support cyclops. Only RadioProcessor
with firmware ID later than 12-15 supports CYCLOPS.
-2 : Wrong number of scans entered. CYCLOPS requires multiple of
4 number of scans.
-3 : No boards found.
|
Creating Additional MATLAB Functions
The functions in the above section were all originally written in C
code, the source files of which are available under the MexSource subdirectory of SPNMR10. The source was
modified from its original form in order that it might be compiled
properly in MATLAB. This modification is as simple as adding a
gateway Mex function that handles the passing of data between the C
code and the MATLAB workspace.
When attempting to compile Mex-files that make use of SpinApi, the
spinapi.h, spinapi.dll and spinapi.lib must all be present in the
current directory for the file to compile properly.
For example, assume I've created a C file called example.c that makes
use of SpinApi that I have formatted to be compiled in MATLAB as a
MEX-file. The command line to compile this file might looks
something as follows:
mex example.c spinapi.lib
Assuming that the example.c and the various spinapi files are located
in the current directory and the compiler has been configured properly,
this should generate a MEX-file with some extension appropriate to the
platform that MATLAB is currently running on.
In order to recompile the MATLAB Interface files (or to compile new
MEX-files) the compiler must
first be configured properly. The following steps explain how to
setup the compiler. This only needs to be done the first time you
compile unless there are settings you want changed:
- Download MinGW and install it to C:\MinGW. You
can dowload MinGW as part of our pre-configured compiler package here.
- Download Cygwin and install it to C:\cygwin.
You will need CygwinDevel.exe. Follow the download and
installation instructions here.
- Download gnumex and extract all files to
C:\gnumex. You can download the gnumex files here.
- Configure the mexopt.bat file by running gnumex.m in
MATLAB. Follow the instructions here.
The following steps need to be done every time you recompile:
- If you wish to download the latest version of the
SpinCore MATLAB interface,
you can find it here.
After it is downloaded, unzip all the files to your working directory.
- Copy the latest versions of spinapi.dll, spinapi.lib
and spinapi.h (and registers.h if necessary) into the MexSource
directory of the unzipped MATLAB files.
- Navigate to that directory in MATLAB and type
"mexCompile.m".
- Now your compilation is complete.
In
addition, the information
located at http://gnumex.sourceforge.net/
may prove to be helpful as well.
Users with later versions of MATLAB may note that the default file type
generated when creating MEX-files is not .dll as the included
precompiled mex-files are for SPNRMR3.
For example, users may find their mex
function generates a .mexw32 file. This is a valid mex-file type
and it should be noted that the .dll was used in the case of this
software to preserve backward compatibility.
© 2010 SpinCore
Technologies |






